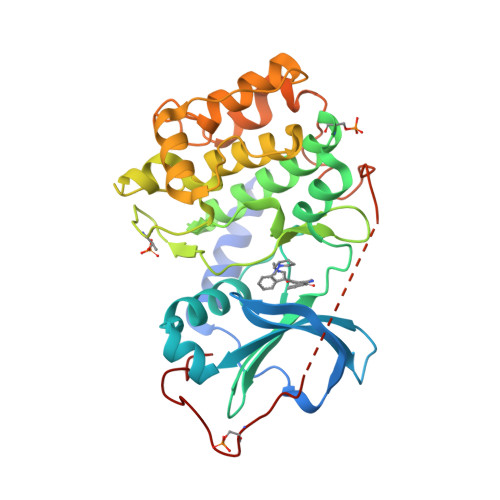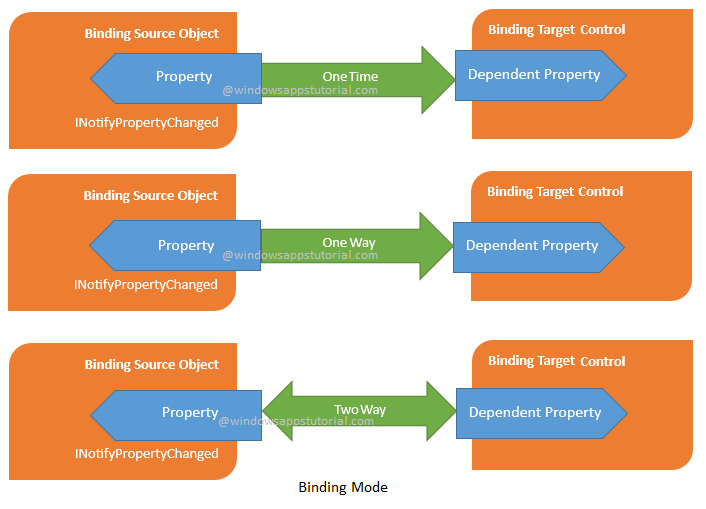
A binding mode hypothesis for prothioconazole binding to CYP51 derived from first principles quantum chemistry | SpringerLink

Binding modes of covalent small-molecule kinase inhibitors (CSKIs). (a)... | Download Scientific Diagram

Binding mode and protein-ligand interactions of 6-((4-fluorophenyl)... | Download Scientific Diagram

Exploration of Type II Binding Mode: A Privileged Approach for Kinase Inhibitor Focused Drug Discovery? | ACS Chemical Biology

Exploration of Type II Binding Mode: A Privileged Approach for Kinase Inhibitor Focused Drug Discovery? | ACS Chemical Biology
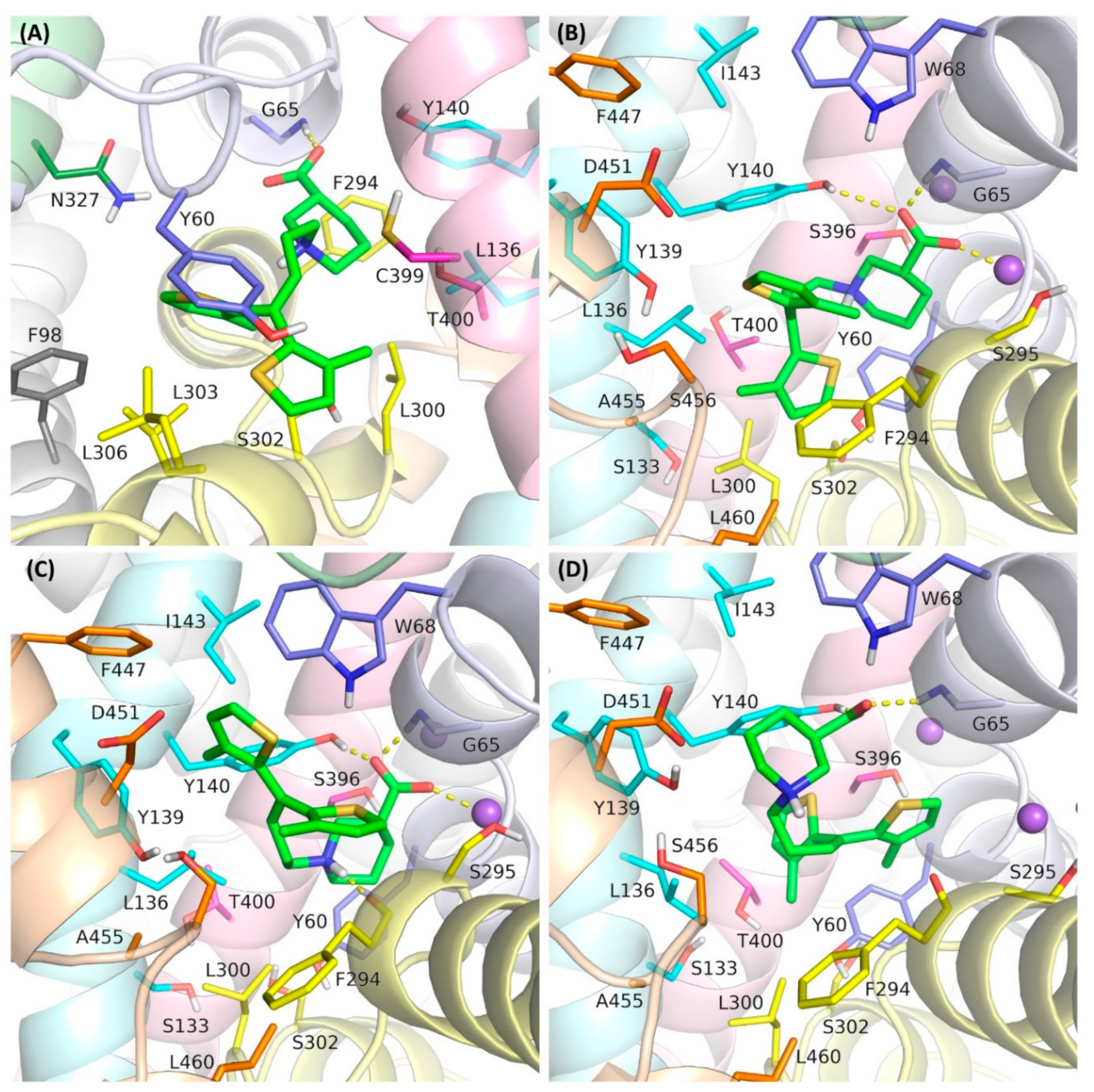
Biomolecules | Free Full-Text | Analysis of Different Binding Modes for Tiagabine within the GAT-1 Transporter
Investigations into the DNA-binding mode of doxorubicinone - Organic & Biomolecular Chemistry (RSC Publishing)

Proposed binding mode of 70551 into the N-Myc-Max DNA-binding domain... | Download Scientific Diagram

(A) The binding mode of 12f (coloured by element with carbons in green)... | Download Scientific Diagram

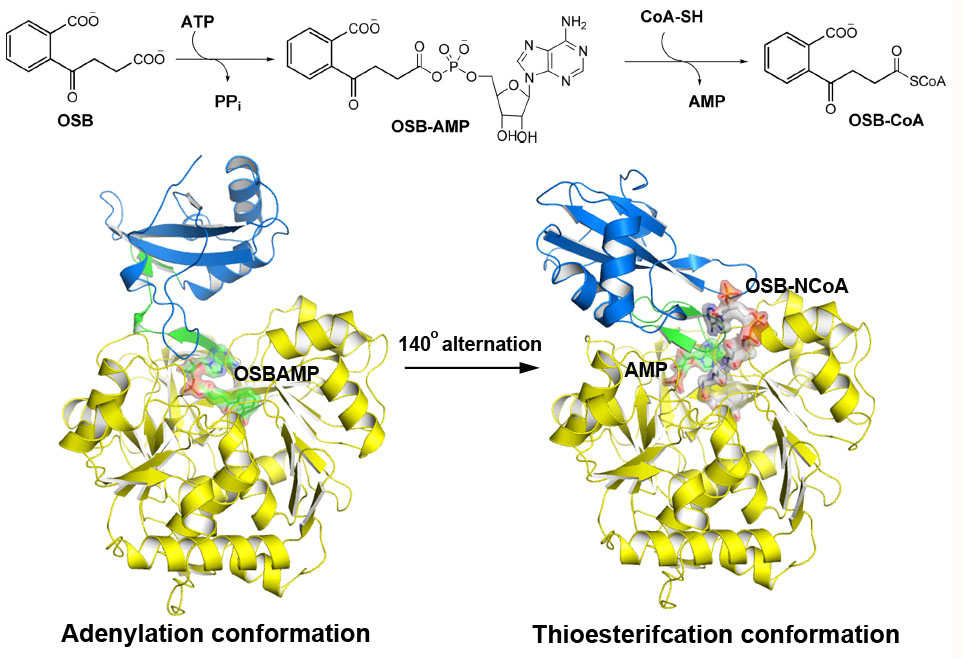
Universität Düsseldorf: Multiple Substrate Binding Mode-Guided Engineering of a Thermophilic PET Hydrolase

Binding mode comparison. (A) Binding mode of compound 10 in PB1(5). (B)... | Download Scientific Diagram

Binding mode of (A) lorlatinib and (B) Hit compound in the wild type... | Download Scientific Diagram
Comparing Binding Modes of Analogous Fragments Using NMR in Fragment-Based Drug Design: Application to PRDX5 | PLOS ONE
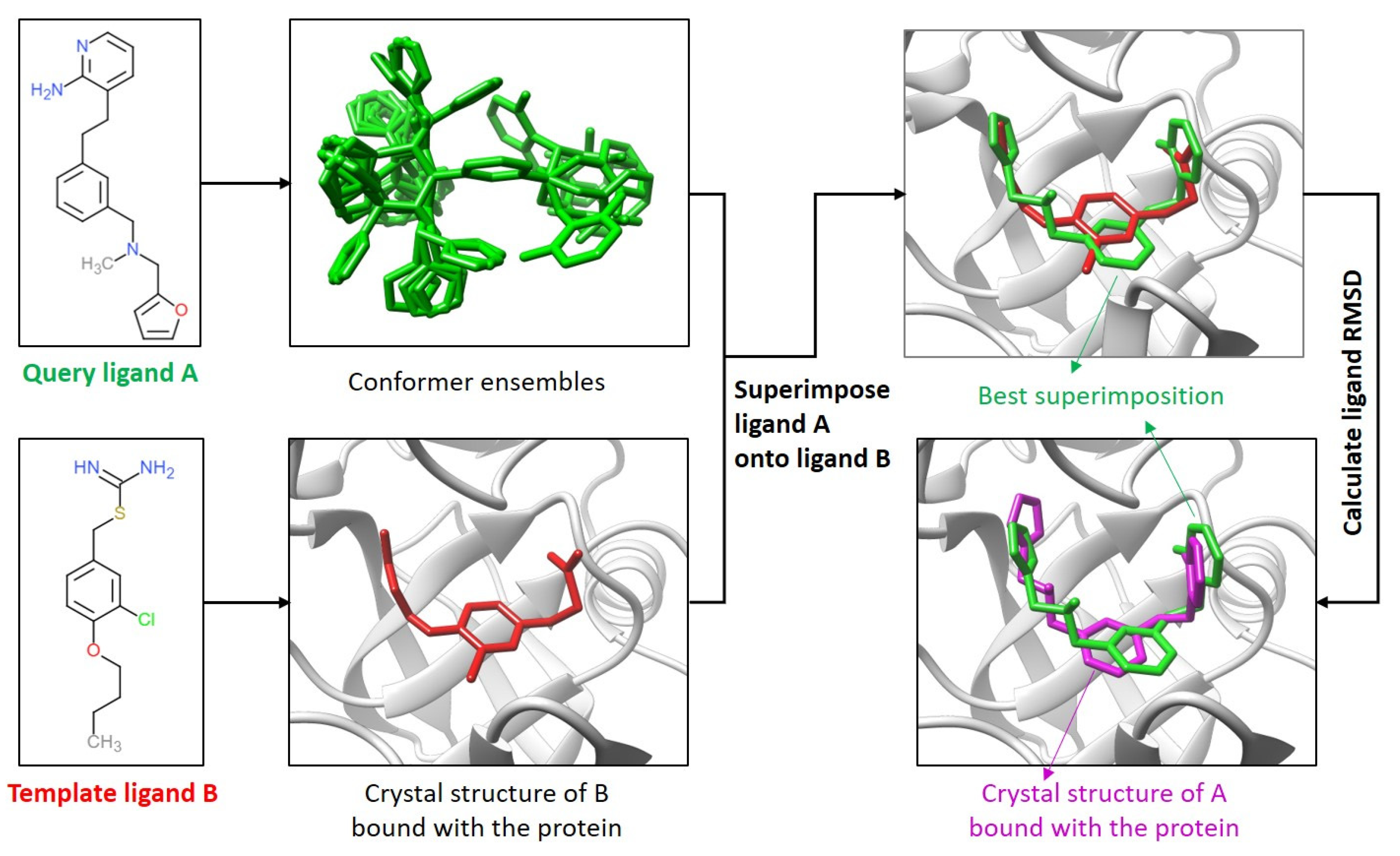
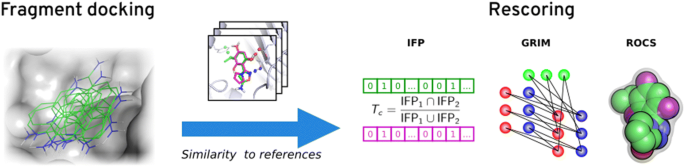
IJMS | Free Full-Text | Dissimilar Ligands Bind in a Similar Fashion: A Guide to Ligand Binding-Mode Prediction with Application to CELPP Studies

Elucidating the Binding Mode between Heparin and Inflammatory Cytokines by Molecular Modeling - Yu - 2021 - ChemistryOpen - Wiley Online Library
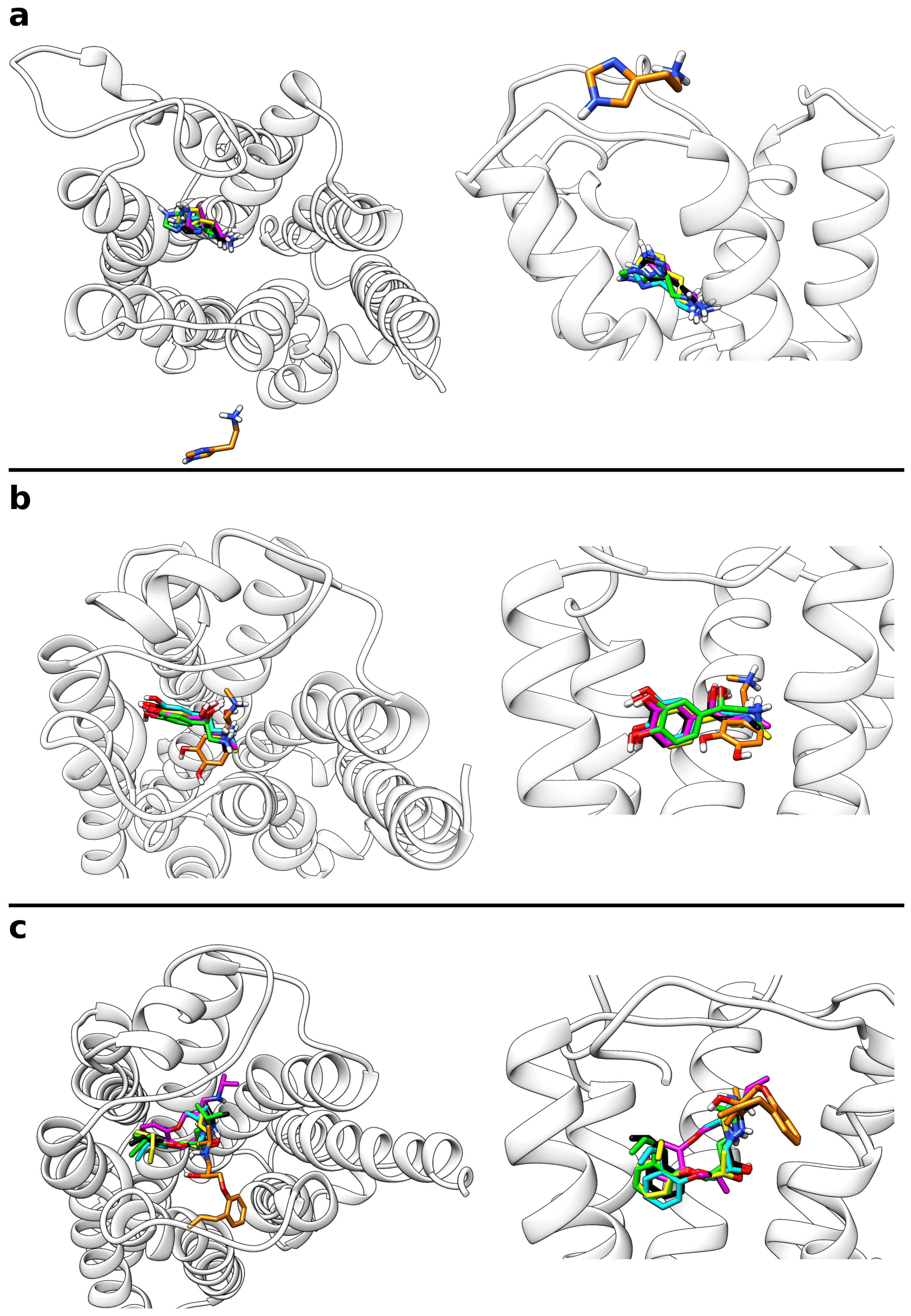
IJMS | Free Full-Text | A Metadynamics-Based Protocol for the Determination of GPCR-Ligand Binding Modes

Erratum: Abl inhibitor BMS354825 binding mode in Ableson kinase revealed by molecular docking studies | Leukemia